Resampling and Feature Engineering
In this tutorial, we will learn about resampling and feature
engineering with the rsample and recipes
packages from tidymodels.
Click the button below to clone the course R tutorials into your DataCamp Workspace. DataCamp Workspace is a free computation environment that allows for execution of R and Python notebooks. Note - you will only have to do this once since all tutorials are included in the DataCamp Workspace GBUS 738 project.
Data Resampling
The first step in fitting a machine learning algorithm involves splitting our data into training and test sets as well as processing our data into a numeric feature matrix.
In machine learning, splitting data into training and test sets is known as resampling, or more generally as cross validation. This is an important step in the model fitting process because it allows us to estimate how our trained machine learning algorithms will perform on new data.
The ultimate goal for any machine learning algorithm is to provide accurate predictions on new, previously unseen data.
Resampling is achieved with the rsample package from
tidymodels. To demonstrate how this done, let’s import the
tidymodels package and the employee_data.
The tidymodels package loads the core machine learning
packages that we will be using this semester, including
rsample, recipes, parsnip,
yardstick, dials, tune, and
workflows. Each one of these packages serves a specific
role in the modeling process. This tutorial will focus on resampling
with rsample and feature engineering with
recipes.
library(tidymodels)── Attaching packages ────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── tidymodels 1.1.0 ──✔ broom 1.0.5 ✔ rsample 1.1.1
✔ dials 1.2.0 ✔ tune 1.1.1
✔ infer 1.0.4 ✔ workflows 1.1.3
✔ modeldata 1.1.0 ✔ workflowsets 1.0.1
✔ parsnip 1.1.0 ✔ yardstick 1.2.0
✔ recipes 1.0.6 ── Conflicts ───────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── tidymodels_conflicts() ──
✖ scales::discard() masks purrr::discard()
✖ dplyr::filter() masks stats::filter()
✖ recipes::fixed() masks stringr::fixed()
✖ kableExtra::group_rows() masks dplyr::group_rows()
✖ dplyr::lag() masks stats::lag()
✖ yardstick::spec() masks readr::spec()
✖ recipes::step() masks stats::step()
• Use tidymodels_prefer() to resolve common conflicts.employee_data <- readRDS(url('https://gmubusinessanalytics.netlify.app/data/employee_data.rds'))
The code below creates a subset of the employee_data
with select columns and a new employee_id variable. This is
so that we can easily demonstrate the use of the recipes
package in the next section.
employee_df <- employee_data %>%
select(left_company, job_level, salary,
weekly_hours, miles_from_home)
# View results
employee_df
Data Splitting
The initial_split() function from the
rsamplepackage is used for generating a data split object
with instructions for randomly assigning rows from a data frame to a
training set and test set. Once the object is created, we can use the
training() and testing() functions to obtain
the two data frames from the object.
When splitting data, it is important to use the
set.seed() function before calling
initial_split(). The set.seed() function takes
any integer as an argument and sets the random number generator in
R to a specific starting point. When this is done, the data
split will be random the first time our code is executed. Every
execution afterwards, will produce the same data split. This guarantees
reproducibility.
The initial_split() function takes three important
arguments, our data, the proportion of rows to add to our training set
(prop), and the variable to use for stratification,
strata.
The default prop value is 0.75. The strata
argument should contain the outcome variable that we are interesting in
predicting. In our case, this is left_company.
Stratification ensures that there are an equal proportion of
left_company values in the training and test sets.
Creating a Data Split Object
First, let’s create a data split object named
employee_split
# Set the random seed
set.seed(314)
employee_split <- initial_split(employee_df, prop = 0.75,
strata = left_company)If we print the employee_split object, we see that we
have 1,103 rows in our training data (known as Analysis in
rsample) and 367 rows in the test data (known as
Assess in rsample)
employee_split<Training/Testing/Total>
<1101/369/1470>
Extracting Training and Test Sets
To create training and test data frames from our
employee_split object, we must pass
employee_split to the training() and
testing() functions.
The code below shows how to do this with the %>%
operator. I have named the resulting data frames
employee_training and employee_test.
When we create the training data, notice that the resulting data frame has 1,103 rows and a random subset of the employees are included.
# Generate a training data frame
employee_training <- employee_split %>% training()
# View results
employee_training
Our test set has 367 rows. Now we are ready to begin our feature engineering steps on the training data.
# Generate a training data frame
employee_test <- employee_split %>% testing()
# View results
employee_test
Feature Engineering
Feature engineering includes all transformations that take a training data set and turn it into a numeric feature matrix.
Typical steps include:
- Scaling and centering numeric predictors
- Removing skewness from numeric variables
- One-hot and dummy variable encoding for categorical variables
- Removing correlated predictors and zero variance variables
- Imputing missing data
Feature engineering steps should be trained on the training data. This includes things such as learning the means and standard deviations to apply in standardizing numeric predictors.
Once these are calculated in the training data, the same transforms are performed on the test data.
This way, the test data is completely removed from the training process and can serve as an independent assessment for model performance.
Specify a Recipe
The first step in build a feature engineering pipeline with the
recipes package is to specify a blueprint for processing
data and assigning roles to each column of the training data.
This is done with the recipe() function. This function
takes two important arguments:
- a model formula
- a data frame or tibble for training the recipe
Model formulas in R have the following form:
outcome ~ predictor_1 + predictor_2 + ...The outcome variable is on the left side of the ~
followed by all predictors separated by a + on the
righthand side.
For example, in our employee_training data, we are
interested in predicting whether an employee will leave the company. Our
response variable is left_company. We would like to use all
other variables as predictors. The way to specify this in an
R formula is as follows:
left_company ~ job_level + salary + weekly_hours + miles_from_homeTypically, model formulas are written using shorthand notation. When
we type left_company ~ ., we are telling R
that left_company is the response variable and all other
variables should be used as predictors.
This saves us from have to type out each predictor variable separated
by a +.
Let’s specify our feature engineering recipe using the
employee_training data and the recipe()
function. We will name our recipe object
employee_recipe
employee_recipe <- recipe(left_company ~ .,
data = employee_training)To explore the variable roles in our recipe, we can pass our recipe
object to the summary() function. This will return a data
frame with 4 columns. The important columns are variable,
type, and role.
The variable column lists all the columns in the input
data, employee_training in this case.
The type column lets us know what data type each
variable has in our training data.
And finally, the role column specifies the various roles
that the recipe() function assigned to each variable based
on our model formula.
Notice that since we used left_company ~ . as our
formula, the left_company variable is assigned as an
outcome variable while all others are assigned as
predictor variables.
summary(employee_recipe)
Processing Numeric Variables
Once we have specified a recipe with a formula, data, and correct
variables roles, we can add data transformation steps with a series of
step() functions. Each step() function in the
recipes package provides functionality for different kinds
of common transformations.
Centering and Scaling
Let’s begin with the simple task of centering and scaling numeric predictor variables. We have been doing this when we subtracted the mean and divided by the standard deviation in our previous tutorials.
The associated step() functions for this task are
step_center() and step_scale(). The
step_center() function subtracts the column mean from a
variable and step_scale() divides by the standard
deviation.
Each successive step() function adds a pre-processing
step to our recipe object in the order that they are
provided.
All step() functions take a recipe as the first
argument, and one or more variables on which to apply the
transformation.
There are special selector functions that can be used to select variables by role (outcome or predictor) or type (numeric or nominal).
all_predictors()- select all predictor columnsall_outcomes()- select the outcome variableall_numeric()- select all numeric columns regardless of roleall_nominal()- select all nominal columns regardless of role
Let’s see what adding these step functions does to our recipe object. We see that we get an updated recipe object as the output with instructions for centering and scaling our numeric columns.
employee_recipe %>%
step_center(salary, weekly_hours, miles_from_home) %>%
step_scale(salary, weekly_hours, miles_from_home)
But how can we obtain the results of the transformations on our
employee_training data frame? We must use the
prep() and bake() functions.
The prep() function trains the recipe on a provided
dataset and the bake() function applies the prepped recipe
to a new data frame of our choice.
Both of these functions take a recipe object as input, so we can
chain the commands with a %>% operator.
The code below takes our employee_recipe, adds centering
and scaling steps on our numeric predictors, trains the steps with
prep() and applies the trained steps to our
employee_test data.
The prep() function has a training argument
which specifies which data to use for training the pre-processing steps,
such as determining the mean and standard deviations of numeric columns
for centering and scaling.
The results from bake() will always be a tibble (data
frame).
employee_recipe %>%
step_center(salary, weekly_hours, miles_from_home) %>%
step_scale(salary, weekly_hours, miles_from_home) %>%
prep(training = employee_training) %>%
bake(new_data = employee_test)
If we wanted to apply our trained recipe to our training data set, it
is as simple as updating the new_data argument in
bake() to a value of NULL . Since the training
data is transformed and a copy is saved during the transformation
process in prep(), NULL instructs the
bake() function to fetch the results.
Passing new_data = employee_training would also work,
but this would re-apply all the transformations to the training data.
For small datasets, this doesn’t make a difference. But for large
datasets, using NULL can save a lot of time.
employee_recipe %>%
step_center(salary, weekly_hours, miles_from_home) %>%
step_scale(salary, weekly_hours, miles_from_home) %>%
prep(training = employee_training) %>%
bake(new_data = NULL)
Using Selector Functions
Instead of specifying the variable names within step()
functions, we can use the special selector functions mentioned
previously.
In this case, we want to center and scale all numeric predictor
variables. We also generally want to exclude processing our outcome
variable. In this case, we don’t have to worry about that since our
response variable is a factor, but it’s good practice to always exclude
the outcome variable with -all_outcomes().
The code below shows how to achieve the previous steps with these special selector functions.
employee_recipe %>%
step_center(all_numeric(), -all_outcomes()) %>%
step_scale(all_numeric(), -all_outcomes()) %>%
prep(training = employee_training) %>%
bake(new_data = employee_test)
step_normalize()
Centering and scaling numeric predictors is so common that there is
one step function, step_normalize(), that does both tasks
at once. The code below takes our employee_recipe, adds a
normalization step to all numeric predictors except the outcome and id
variables, and applies the trained recipe to the
employee_test data.
Notice that we get the same results as above.
employee_recipe %>%
step_normalize(all_numeric(), -all_outcomes()) %>%
prep(training = employee_training) %>%
bake(new_data = employee_test)
Transforming Highly Skewed Data
The step_YeoJohnson() function is used to removing
skewness in numeric data. This is a special transformation that tries to
map the original values of a numeric variable to the normal
distribution.
Before we use this function, lets have a look at the distribution of
the miles_from_home variable in
employee_training.
Note: Another common method for dealing with skewed data is to apply a logarithm transform (usually to base 10). This, however, requires that all numeric data values are greater than zero. The Yeo-Johnson transformation will work on numeric variables that have zero or negative values.
ggplot(data = employee_training, mapping = aes(x = miles_from_home)) +
geom_histogram(fill = '#006EA1', color = 'white', bins = 10) +
labs(title = 'Distribution of Miles From Home',
x = 'Miles from Home',
y = 'Number of Employees')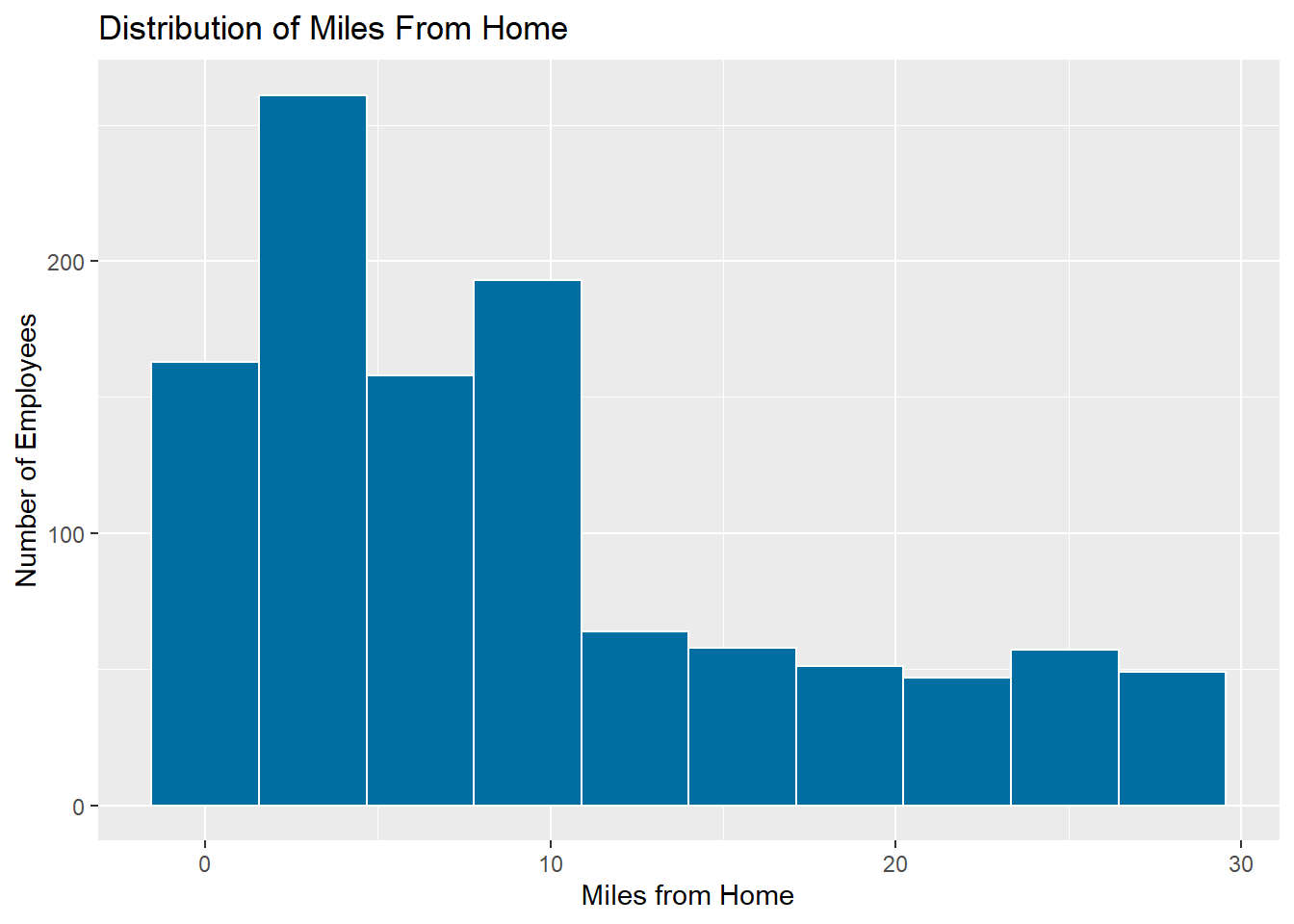
Now let’s transform this variable in the training data with the Yeo-Johnson transformation and look at the resulting values.
employee_recipe %>%
step_YeoJohnson(miles_from_home) %>%
prep(training = employee_training) %>%
bake(new_data = NULL)
Let’s plot the distribution of the results. In the code below, I pipe
the results from above into ggplot. Although the results
are not perfectly symmetric, they are much better than the original
distribution of values. In general, I recommend performing this step on
all numeric predictors.
employee_recipe %>%
step_YeoJohnson(miles_from_home) %>%
prep(training = employee_training) %>%
bake(new_data = NULL) %>%
ggplot(mapping = aes(x = miles_from_home)) +
geom_histogram(fill = '#006EA1', color = 'white', bins = 10) +
labs(title = 'Distribution of Transformed Miles From Home',
x = 'Miles from Home',
y = 'Number of Employees')
Putting It All Together
To demonstrate how a full recipe is specified, let’s create a recipe
object called employee_numeric that will train the
following steps on our employee_training data:
- remove highly correlated predictors
- remove skewness from all numeric predictors
- center and scale all numeric predictors
employee_numeric <- recipe(left_company ~ .,
data = employee_training) %>%
step_corr(all_numeric(), -all_outcomes()) %>%
step_YeoJohnson(all_numeric(), -all_outcomes()) %>%
step_normalize(all_numeric(), -all_outcomes()) %>%
prep(training = employee_training)Now that we have our trained recipe, we can apply the transformations
to our training and test data with bake().
processed_employee_training <- employee_numeric %>%
bake(new_data = NULL)
processed_employee_test <- employee_numeric %>%
bake(new_data = employee_test)# View results
processed_employee_training# View results
processed_employee_test
Processing Categorical Variables
For a large number of machine learning algorithms, all data in a feature matrix must be numeric. Therefore, any character or factor variables in a data frame must be transformed into numbers.
How is this done? The two primary methods are dummy variable creation
and one-hot encoding. Both methods are performed by the
step_dummy() function.
Let’s see an example of both methods using our
employee_recipe object. We will transform the
job_level variable using one-hot encoding and dummy
variables.
One-Hot Encoding
The job_level variable in employee_training
has 5 unique values: Associate, Manager, Senior Manager, Director, and
Vice President.
One-hot encoding will produce 5 new variables that are either 0 or 1
depending on whether the value was present in the job_level
row.
The new variables are created with the following naming convention:
variable_name_level
For example, job_level_Associate will be one variable
that is created. If the value of job_level for any row in
the data is “Associate”, then this new variable will be equal to 1 and 0
otherwise.
Let’s see how we can do this with step_dummy()
# One-hot encode job_level
employee_recipe %>%
step_dummy(job_level, one_hot = TRUE) %>%
prep(training = employee_training) %>%
bake(new_data = NULL)
Dummy Variables
Creating dummy variables is similar to one-hot encoding, except that
one level is always left out. Therefore, if we create dummy variables
from the job_level() function, we will have 4 new variables
instead of 5.
This method is generally preferred to one-hot encoding because many statistical models will fail with one-hot encoding. This is because it creates multicollinearity in the one-hot encoded variables.
In fact, the default of step_dummy() is to have
one_hot set to FALSE. This is what I recommend for most
machine learning applications.
Let’s see the difference when we use the default settings of
step_dummy(). Notice that job_level_Associate
is now excluded.
employee_recipe %>%
step_dummy(job_level) %>%
prep(training = employee_training) %>%
bake(new_data = NULL)
Creating a Feature Engineering Pipeline
When creating feature engineering recipes with many steps, we have to keep in mind that the transformations are carried out in the order that we enter them.
So if we use step_dummy() before
step_normalize() our dummy variables will be normalized
because they are numeric at the point when step_normalize()
is called.
To make sure we don’t get any unexpected results, it’s best to use the following ordering of high-level transformations:
- Correlation filters and skewness transformations -
step_YeoJohnson()andstep_corr() - Centering, scaling, or normalization on numeric predictors
- Dummy variables for categorical data
Let’s put together all that we have learned to create the following
feature engineering pipeline on the employee_training
data:
- Correct for skewness on all numeric predictors
- Normalize all numeric predictors
- Create dummy variables for all character or factor predictors (we can use the all_nominal() selector for this)
employee_transformations <- recipe(left_company ~ .,
data = employee_training) %>%
# Transformation steps
step_YeoJohnson(all_numeric(), -all_outcomes()) %>%
step_normalize(all_numeric(), -all_outcomes()) %>%
step_dummy(all_nominal(), -all_outcomes()) %>%
# Train transformations on employee_training
prep(training = employee_training)
# Apply to employee_test
employee_transformations %>%
bake(new_data = employee_test)Copyright © David Svancer 2023 |
